A Short Primer on Y-Marker Analysis
© 2008 James M. Gossett, Ithaca, NY
All materials contained on this site are protected by United States copyright law and may not be reproduced, distributed, transmitted, displayed, published or broadcast without the prior written permission of the authors. However, you may download material for your personal, noncommercial use only.
Y-chromosomal DNA (YDNA) is passed from father to son, from son to grandson, from grandson to great-grandson, etc. -- usually unchanged in sequence. However, occasionally a random mutation occurs in the replication process when a son is produced, introducing a change in the YDNA sequence of nucleic-acid bases that this son will, in turn, pass along to his son. Tracking such changes in specific regions ("markers") of the YDNA is the basis of YDNA genealogical analysis. Only males possess the Y-chromosome; therefore, YDNA-based genealogical analysis is uniquely suited for surname studies, since sons generally inherit their fathers' surnames, along with their Y-chromosomes.
This is a short primer on how Y-chromosome markers are measured. Much has been greatly simplified – though I hope not overly so.
Background
The DNA code is designated by a sequence of nucleic-acid bases. There are only four bases – guanine (G), cytosine (C), adenine (A) and thymine (T). The DNA molecule is actually a double-stranded helix, held together with complementary pairing of bases across the two strands. G bonds only to C; and A bonds only to T.
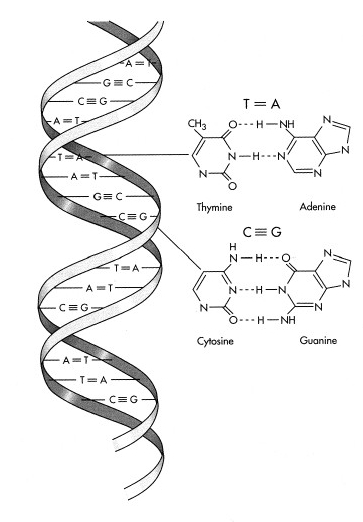
http://www.ucmp.berkeley.edu/glossary/gloss3/dna.html
Markers used in Male-Male Lineage Tracing
Markers on the Y chromosome, used to track genealogical lineage, are areas of “junk” DNA that don’t encode for anything functional, but consist of short tandem repeats (STRs) – short sequences of DNA bases repeated a number of times. The number of times the short sequence is repeated at the particular marker site is called its “allele” length. Since these STRs don’t encode for anything important, mutations or changes in their sequence lengths don’t affect survival and are faithfully transmitted to sons thereafter – until the next mutation event causes yet another change in this or some other marker’s allele length. Accumulated changes in allele length of various Y-markers are thus used to track lineage.
Here’s an example of a marker that has a STR of the bases “TAG” repeated four times – i.e., an allele length of 4.
3’ 5’
..-T-A-A-C-G-G-T-A-G-T-A-G-T-A-G-T-A-G-A-A-T-G-....
a STR of “TAG” repeated 4 times
Since DNA is double-stranded, there is a complementary strand paired with it, in the intact chromosome:
5’ 3’
…..-A-T-T-G-C-C-A-T-C-A-T-C-A-T-C-A-T-C-T-T-A-C-……
a STR of “ATC” repeated 4 times
[Note: the designations, 3’ and 5’, just refer to which end of the DNA molecule is which.]
Thus, if you wanted to, you could have referred to this STR as either a 4-fold repeat of “TAG” or a 4-fold repeat of “ATC” – depending upon which of the two complementary strands you chose to reference. There is a standard convention, but that need not concern us here.
Marker Analysis
Using a technique known as polymerase chain reaction (PCR), the STR can be replicated millions of times to increase concentration to measurable levels. Biologists refer to this many-fold replication as “amplification,” and the amplified segment is called the “amplicon.” The PCR reaction requires a “primer pair” – which are relatively short (but unique) sequences that are complementary to sequences that flank (perhaps even including part of) the STR. When double-stranded DNA is heated, the two strands separate, allowing replication. The primer pair is added in many-fold excess, along with some special enzymes (that allow synthesis of new complementary strands to be paired – or “hybridized” -- to the original strands) and large concentrations of DNA backbone molecules of each base-type (G, C. A, and T). The PCR reaction occurs, creating a copies of the STR-portions that are complementary to the originals (plus flanking primers):
3’ 5’
….-T-A-A-C-G-G-T-A-G-T-A-G-T-A-G-T-A-G-A-A-T-G-…..
T-G-C-C-->
forward primer
G C G C
C
G T A T
C G C A
C
G T A T
C G C A
T C T reverse primer
5’ <--A-A-T-G 3’
…..-A-T-T-G-C-C-A-T-C-A-T-C-A-T-C-A-T-C-T-T-A-C-……
Then:
3’ 5’
…..-T-A-A-C-G-G-T-A-G-T-A-G-T-A-G-T-A-G-A-A-T-G-…..
T-G-C-C-A-T-C-A-T-C-A-T-C-A-T-C-T-T-A-C
\
amplicons
\
5’ A-C-G-G-T-A-G-T-A-G-T-A-G-T-A-G-A-A-T-G 3’
…..-A-T-T-G-C-C-A-T-C-A-T-C-A-T-C-A-T-C-T-T-A-C-……
Both complementary amplicons are of the same length. With each cycle of the PCR reaction, the number of copies of amplicons is doubled. The PCR reaction is allowed to cyclically repeat – again and again. So, after one cycle there are 2 copies; after two cycles, 4 copies; ..... after 30 cycles there are 2 to the 30th power (or about a billion) copies.
The purpose of PCR is to selectively amplify sequences of interest. After so-doing, the reaction product is analyzed by capillary electrophoresis (CE) that allows the length of the amplicon to be determined. Put very simply, CE uses a very fine capillary tube packed with a gel (sort of like a high-tech Jello®) with a high voltage applied across its length. The voltage is applied at a very high magnitude (ca. 3000 volts) for a relatively brief duration (ca. 10 seconds). The DNA amplicon has a charge, and it will therefore migrate down the capillary, attracted by the opposite polarity at the other end. The smaller the amplicon, the faster is migrates – and hence the further it will have migrated down the capillary during the duration of applied voltage. Thus, CE separates amplicons by size. CE is very sensitive and can discriminate between two amplicons that are different in length by only one nucleic-acid base! The primers used in PCR are typically tagged with a molecule that causes them to fluoresce, and that’s how the resulting positions of the amplicons in the capillary are detected in CE. Standards of different amplicon lengths are run to calibrate the CE so that position can be matched precisely to amplicon length.
So, suppose Jeff has an allele length of four for this particular marker; and Jim has an allele length of only three for the same marker. This difference will manifest itself as Jeff’s amplicon being three bases longer than Jim’s. Such a difference in allele length is readily detectible with CE. On a Y-marker test, Jeff’s result for this particular marker will be reported as an allele length of “4”, whereas Jim’s result will be “3”.
Multiplex Analysis
For high-throughput, automated analysis (such as what FTDNA is doing), the type of analysis I’ve described above can be done with a “panel” of multiple markers simultaneously – i.e., in “multiplex.”
FTDNA performs analyses with panels of from 7 to 13 markers at once. In principle it’s doable at least up to 20 markers simultaneously. Suppose that, instead of using a single primer pair-type, you simultaneously use primer pairs for each of 12 markers. You could run a Y12 test – the full “panel” of markers – all at once. The different primer pairs could be tagged with different fluorescent molecules, to aid in discriminating the different amplicons in the CE assay. Add all 12 primer pairs (appropriately fluorescently tagged); run the PCR through sufficient number of cycles to get detectable concentrations of all amplicons; then assay the PCR product by CE to separate the amplicons and determine the allele length of each marker.
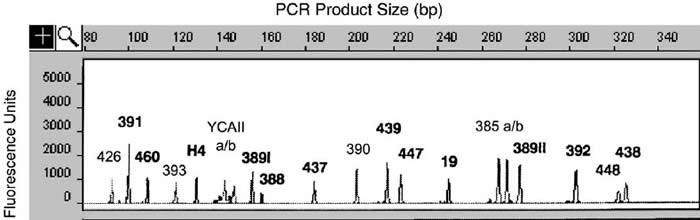
An example of a run with 20 simultaneous markers is shown below.
From: Butler, J. M. et al., “A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers,” Forensic Science International 129, 10-24 (2002).
FTDNA apparently performs multiplex analyses of from 7 to 13 markers per individual per reaction – the number per reaction depending on peculiarities of specific markers (some may mutually interfere with each other, primer-wise, or in terms of CE resolution). That’s why FTDNA reports results of tests in marker-groupings such as “YDNA12; YDNA13-25; YDNA26-37; YDNA38-47; YDNA 48-60; YDNA 61-67.”
Quality-Control Issues
Where are the places where things can go awry? These four spring to mind: (1) human error in sample attribution (i.e., mis-attributing a sample result to the wrong client); (2) bad primers; (3) contaminated reagents; and (4) inaccurate calibration of CE.
Of these, only (1) is likely to go undetected. (2) and (3) cause generally catastrophic errors (either no PCR amplification or false positives, respectively). (4) is readily detected by running standards or controls along with samples -- those would be the ‘cross check’ controls described by Dr. Thomas Krahn in his response to an inquiry about quality control:
“In every panel we have control markers that will be run twice in different panels. The control markers are analyzed with different primers, so that we really have an independent control. E.g.: the first 12 (14) markers are not run in a single multiplex, but they contain two markers that are present in both multiplexes. Those two markers serve as crosscheck controls. Only if they match, we consider an analysis as confirmed and only then we return the results to the customer.”
I can’t say what steps FTDNA takes to prevent human errors in sample attribution, but I’m confident they have paid attention to that potential problem, too.